Green Fluorescent Protein
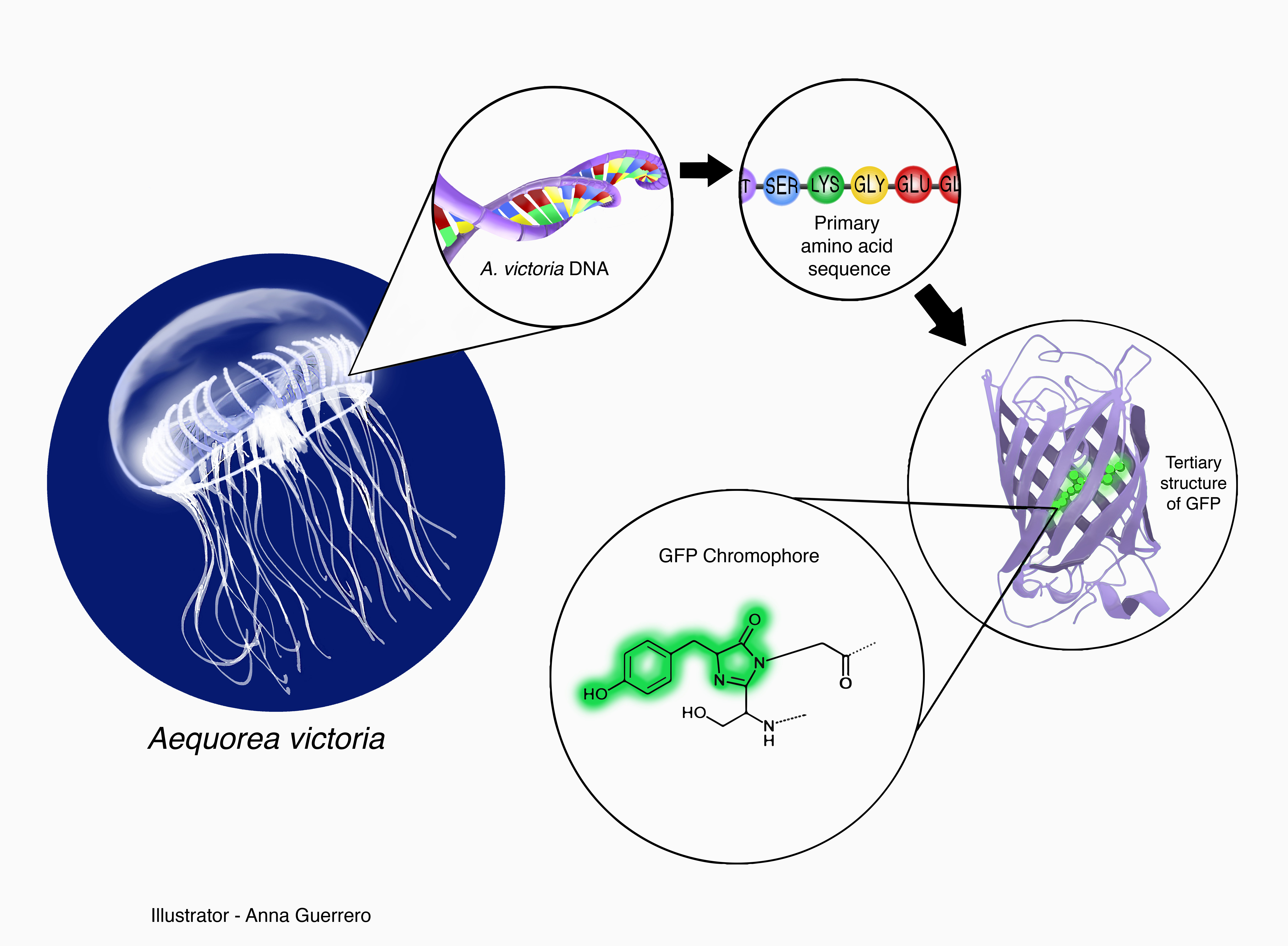 Editor's note: Anna Guerrero created the above image for this article. You can find the full image and all relevant information here.
Editor's note: Anna Guerrero created the above image for this article. You can find the full image and all relevant information here.
Green fluorescent protein (GFP) is a protein in the jellyfish Aequorea Victoria that exhibits green fluorescence when exposed to light. The protein has 238 amino acids, three of them (Numbers 65 to 67) form a structure that emits visible green fluorescent light. In the jellyfish, GFP interacts with another protein, called aequorin, which emits blue light when added with calcium. Biologists use GFP to study cells in embryos and fetuses during developmental processes.
Biologists use GFP as a marker protein. GFP can attach to and mark another protein with fluorescence, enabling scientists to see the presence of the particular protein in an organic structure. Gfp refers to the gene that produces green fluorescent protein. Using DNA recombinant technology, scientists combine the Gfp gene to a another gene that produces a protein that they want to study, and then they insert the complex into a cell. If the cell produces the green fluorescence, scientists infer that the cell expresses the target gene as well. Moreover, scientists use GFP to label specific organelles, cells, tissues. As the Gfp gene is heritable, the descendants of labeled entities also exhibit green fluorescence.
Edmund N. Harvey, a professor at Princeton University in Princeton, New Jersey, initiated the studies on bioluminescence in the US. In 1921, Harvey described the yellow tissues in the umbrella of jellyfish as being luminous in particular conditions, such as at night or when the jellyfish is stimulated with electricity. In 1955, Demorest Davenport at the University of California at Santa Barbara in Santa Barbara, California, and Joseph Nicol at Plymouth Marine Laboratory in Plymouth, England, used photoelectric recording and histological methods to confirm Harvey's descriptions, and they identified the green fluorescent materials in the marginal canal of the umbrella.
In the same year, Osamu Shimomura became a research assistant at Nagoya University in Nagoya, Japan, and he crystallized the luciferin, a light-emitting compound found in the sea-firefly Vargula hilgendorfii. Shimomura published his results in 1957. One of Harvey's students, Frank H. Johnson, studied bioluminescence at Princeton University. Johnson followed Shimomura's work and invited him to work in the US, and in 1960 Shimomura received a Fulbright Travel Grant and started working with Johnson. Shortly after Shimomura arrived in the US, Johnson introduced the bioluminescence of Aequorea Victoria to Shimomura. In the US, jellyfish live only on the west coast, so Shimomura traveled to the Friday Harbor Laboratories of the University of Washington in San Juan Island, Washington, during the summer of 1961. After catching about 10,000 jellyfish, Shimomura took the extracts of the jellyfish and preserved it in dry-ice to bring it back to Princeton in September of 1961.
At Princeton, Shimomura and his colleagues started to purify the bioluminescent substance, and they found that it was a protein, which they called aequorin. When they purified aequorin, they also discovered traces of another protein, which showed green fluorescence. Shimomura's team published the findings in "Exraction, Purification, and Properties of Aequorin" in 1962. The paper was about aequorin, but it also described a green protein, which exhibited green fluorescence under sunlight. John W. Hasting and James G. Morin, who later researched aequorin, termed the protein as green fluorescent protein in 1971.
Shimomura focused on aequorin, purified the protein, crystallized it, and elucidated its underlying structure. He also studied the properties of GFP, and published his last paper on GFP in 1979. In 1981, after leaving Princeton University for the Marine Biology Laboratory in Woods Hole, Massachusetts, Shimomura did not research on GFP anymore. From 1979 to 1992, many researchers studied various aspects of GFP, including the use of Nuclear Magnetic Resonance to study the amino acids of the protein, the use of X-rays to study its crystal, and the evolution of GFP.
In the early 1990s, molecular biologist Douglas Prasher, at the Marine Biology Laboratory, used GFP to design probes, a technology involving fragments of DNA to detect the presence of nucleotide sequences. Prasher isolated the complementary DNA (cDNA) of Gfp gene, and he published the sequence of the gene in 1992. After the publication of the cDNA sequence in 1992, Prasher's funding from the American Cancer Society in Atlanta, Georgia, expired. When he applied for funding from the US National Institute of Health in Bethesda, Maryland, the reviewer argued that Prasher's research lacked contributions to society. As Prasher could not secure funding to support his research any further, he left the Marine Biology Laboratory to work for the US Department of Agriculture in Massachusetts.
After Prasher's publication in 1992, many scientists tried to transfer and express the Gfp gene in organisms other than jellyfish using DNA recombinant technology, and Martin Chalfie was the first who succeeded. Chalfie, a Professor at Columbia University in New York, New York, studied the development of the nematode Caenorhabditis elegans. Chalfie heard about the protein GFP in a lecture, and he speculated that GFP might facilitate his study of gene expression in C. elegans. Chalfie's team obtained the cDNA of the gene Gfp from Prasher and inserted only the coding sequence of Gfp gene first in the bacterium Escherichia Coli, and then in C. elegans. Chalfie and his team found that Gfp gene produced GFP without added enzymes or substrates in both organisms. In 1994, Chalfie published his results in "Green Fluorescent Protein as a Marker for Gene Expression". The detection of GFP needed only ultraviolet light. Thereafter, many biologists introduced GFP into their experiments to study gene expression. Satoshi Inouye and Frederick Tsuji at Princeton University also expressed Gfp in E. Coli in 1994.
Many scientists tried to mutate the Gfp gene to make the resultant protein react to wider wavelengths and emanate different colors. Other scientists studied different fluorescent proteins (FPs). Roger Tsien, a professor at the University of California San Diego, in San Diego, California, reengineered the gene Gfp to produce the protein in different structures. His team also reengineered other FPs. Due to Tsien's and other bioengineers' efforts, GFP could not only exhibit brighter fluorescence, but also respond to a wider range of wavelengths, as well as emit almost all colors, except for red. Tsien's findings enabled scientists to tag multiple colored GFPs to different proteins, cells, or organelles of interest, and scientists could study the interaction of those particles. Red FP became available in 1999, when Sergey Lukyanov's team at the Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry in Moscow, Russia, found that some corals contained the red fluorescent protein, called DsRed. Other laboratories developed fluorescent sensors for calcium, protease and other biological molecules. Since then, scientists have reported more than 150 distinct GFP-like proteins in many species.
As GFP does not interfere with biological processes when used in vivo, biologists use it to study how organisms develop. For example, after 1994, Chalfie and his colleagues applied GFP in the study of the neuron development of C. elegans. In a 2002 paper, Chalfie and his colleagues describe how they first labeled a specific gene involved in tactile perception in neuron cells with GFP, and then observed the amount of fluorescence emitted by those cells. Because mutant cells produced less or more GFP than normal cells, the abnormal amount of fluorescence production indicated the abnormal development of mutants. Since then, this field of research expanded to many other organisms, including fruitflies, mice, and zebra fish.
On 10 December 2008, The Royal Swedish Academy of Science academy awarded the Noble Prize in Chemistry to Tsien, Chalfie, and Shimomura for their discoveries on GFP.
Sources
- Chalfie, Martin, Yuan Tu, Ghia Euskirchen, William W. Ward, and Douglas C. Prasher. "Green Fluorescent Protein as a Marker for Gene Expression." Science 263 (1994): 802–05.
- Chalfie, Martin. "GFP: Lighting Up Life (Nobel Lecture)." http://www.nobelprize.org/nobel_prizes/ chemistry/laureates/2008/ chalfie_lecture.pdf (Accessed February 12, 2014).
- Davenport, Demorest and Joseph Nicol. "Luminescence of Hydromedusae." Proceedings of the Royal Society B: Biological Sciences 144 (1955): 399–11.
- Harvey, Edmund. "Studies on bioluminescence. XIII. Luminescence in the coelenterates." Biological Bulletin 41 (1921): 280–87. http://archive. org/details/jstor-1536528 (Accessed February 21, 2014).
- Hastings, John, and James Morin. "Comparative Biochemistry of Calcium Activated Photo Proteins from the Ctenophore, Mnemiopsis and the Coelenterates Aequorea, Obelia, Pelagia and Renilla." The Biological Bulletin 137 (1969): 402. http:/ /www.biolbull.org/content/137/2/384.full.pdf (Accessed February 11, 2014).
- Matz, Mikhail V., Arkady F. Fradkov, Yulii A. Labas, Aleksandr P. Savitsky, Andrey G. Zaraisky, Mikhail L. Markelov, and Sergey A. Lukyanov. "Fluorescent proteins from nonbioluminescent Anthozoa species." Nature Biotechnology 17 (1999): 969–73.
- Nienhaus, Ulrich. "The Green Fluorescent Protein: A Key Tool to Study Chemical Processes in Living Cells." Angewandte Chemie International Edition 47 (2008): 8992–94.
- "Osamu Shimomura – Biographical". http://www.nobelprize.org/nobel_prizes/ chemistry/laureates/2008/ shimomura-bio.html (Accessed February 12, 2014).
- Prasher, Douglas C, Virgina K. Eckenrodeb, William W. Ward, Frank G. Prendergastd, and Milton J. Cormier. "Primary Structure of the Aequorea Victoria Green-Fluorescent Protein." Gene 111 (1992): 229–33.
- Shimomura, Osamu, Frank H. Johnson, and Yo Saiga. "Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from Luminous Hydromedusan Aequorea." Journal of Cellular and Comparative Physiology 59 (1962): 223–39.
- Shimomura, Osamu. "Structure of the Chromophore of Aequorea Green Fluorescent Protein." FEBS Letter 104 (1979): 220–22. http://dx. doi.org/10.1016/0014-5793(79)80818-2 (Accessed February 21, 2014).
- Shimomura, Osamu. "The Discovery of Aequorin and Green Fluorescent Protein." Journal of Microscopy 217 (2005): 3–15. "The Nobel Prize in Chemistry 2008". http://www.nobelprize.org/nobel_prizes/chemistry/ laureates/2008/ (Accessed February 12, 2014).
- Tsien, Roger Y. "The Green Fluorescent Protein." Annual Review of Biochemistry 67 (1998): 509–44.
- Tsuji, Frederick. "Early History, Discovery, and Expression of Aequorea Green Fluorescent Protein, with a Note on an Unfinished Experiment." Microscopy Research and Technique 73 (2010): 785–96.
- Zhang, Yun, Charles Ma, Thomas Delohery, Brian Nasipak, Barrett C. Foat, Alexander Bounoutas, Harmen J. Bussemaker, Stuart K. Kim, and Martin Chalfie. "Identification of genes expressed in C. elegans touch receptor neurons." Nature 418 (2002): 331–35.
- Zimmer, Mark. "GFP: from Jellyfish to the Nobel Prize and Beyond." Chemical Society Reviews 38 (2009): 2823–32. http://pubs.rsc.org/en/content/articlepdf/2009/cs/b904023d (Accessed February 12, 2014).
Keywords
Editor
How to cite
Publisher
Handle
Rights
Articles Rights and Graphics
Copyright Arizona Board of Regents Licensed as Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported (CC BY-NC-SA 3.0)